Welcome to the INFERNET project:
new algorithms for inference and optimization from large-scale biological data
Transfer ideas from statistical inference, optimization techniques and high-performance computing methods
into the world of quantitative biology
A consortium characterized by proven track-record of high-quality research





The project relies on a secondments plan to enhance the potential and future career perspectives of the staff members
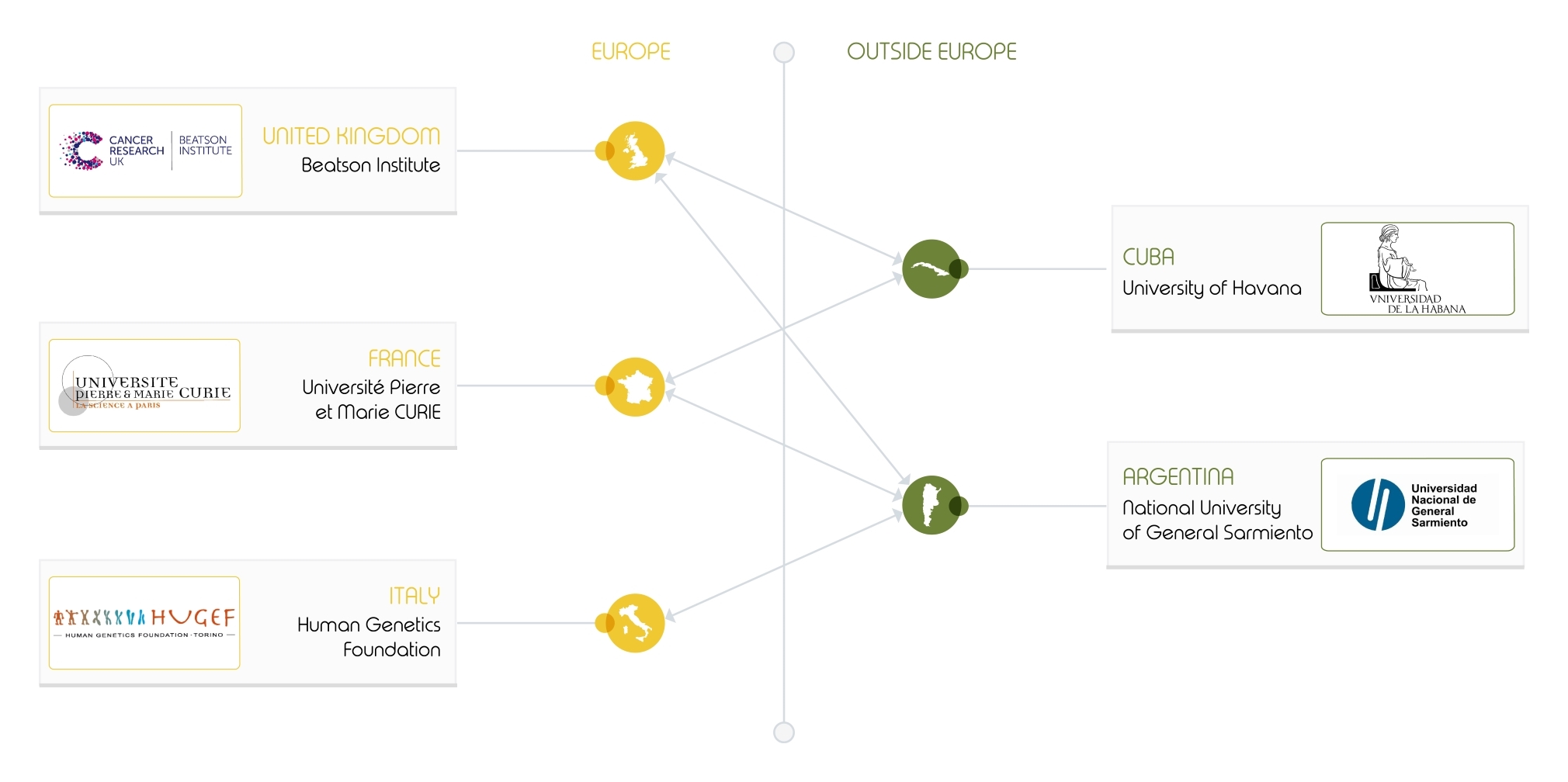
The INFERNET project in a nutshell
Implementing a highly integrated research program leading from the design of new algorithms to concrete biological applications
Two main research themes will be covered by the consortium
Research Field
INFERENCE OF INTERACTION NETWORKS FROM DATA
A major challenge in computational biology is to use data to unveil the interrelations between biological processes and the molecules contributing to them in terms of regulatory networks. The analysis will produce topological description (who is directly coupled with whom?), and quantitative functional description (how things interact?).
Research Field
ANALYSIS OF STATIC AND DYNAMICAL PROCESSES ON NETWORKS
INFERNET aim at developing distributed algorithmic techniques for a few key inference problems in molecular systems biology such as large scale models of proliferative metabolism, and large scale models of post-transcriptional microRNA mediated regulation.
Application domains can be broken down into four main areas
Application Domain
INFERENCE AND MODELING OF MULTI-SCALE BIOLOGICAL NETWORKS
INFERNET research will be applied to inference from scarce, noisy data sets where a central question is the design of adequate null models to assign statistical significance values to the inferred model parameters, to the sparse graphical model learning, and to the taming with the algorithmic complexity.
Application Domain
RATIONAL DESIGN OF BIOLOGICAL MOLECULES
The project will exploit the co-evolutionary information gathered either from existing databases of thousands of functionally active mutants, to build reliable multivariate models of the observed sequence variability to predict new functional molecules.
Application Domain
QUANTITATIVE STUDY OF CELL ENERGETICS IN PROLIFERATIVE REGIMES
Constraint-based models are powerful mathematical tools that are used to examine metabolic networks at genome scale, and in particular to formulate testable hypotheses on aberrant regimes such as proliferative cancer.
Application Domain
FUNCTIONAL STATES OF LARGE-SCALE REGULATORY NETWORKS
Characterization of regulatory mechanisms in post-transcriptional microRNA mediated networks to: (i) unveil their physical origin, magnitude and dependence on kinetic parameters and (ii) under which conditions it performs other regulatory mechanisms.

New algorithms for inference and optimization from large-scale biological data
MEET INFERNET
FOLLOW US

The INFERNET project is co-funded by the European Union’s H2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement number 734439
